A Parasite Constantly Changes Disguises to Survive
To survive, a parasite mixes and matches its disguises, study suggests
When hiding out in blood, sleeping sickness parasites must get creative with their repertoire of camouflage genes
Orchestrated costume changes make it possible for certain nasty microbes to outsmart the immune system, which would otherwise recognize them by the telltale proteins they wear. By taking the first detailed look at how one such parasite periodically assumes a new protein disguise during a long-term infection, new research at Rockefeller University challenges many assumptions about one of the best-known examples of this strategy, called antigenic variation, in the parasite that causes African sleeping sickness.
Widget not in any sidebars
Watch video: http://phys.org/news/2015-03-survive-parasite-disguises.html
For research published on March 27 in Science, a team at Rockefeller University tracked the appearance and disappearance of these protein coats within the blood of mice infected by Trypanosoma brucei. Their results reveal an unexpected diversity in disguises present at any one time and challenge the conventional understanding of the dynamics that allow the parasite to persist.
Spread by the Tsetse fly in rural Africa, T. brucei travels through the blood and across the blood-brain barrier to cause potentially fatal disease that disrupts circadian rhythms, hence the name sleeping sickness. T. brucei, like the malaria parasite and some other pathogens, relies on antigenic variation to stay one step ahead of their hosts’ immune systems.
Here’s how it works. Many animals, including humans, have immune systems capable of learning to recognize pathogens based on those pathogens’ antigens, usually proteins on their surface. After encountering an antigen, the immune system generates its own proteins called antibodies to target that antigen. By continually changing antigens, a pathogen evades those antibodies.
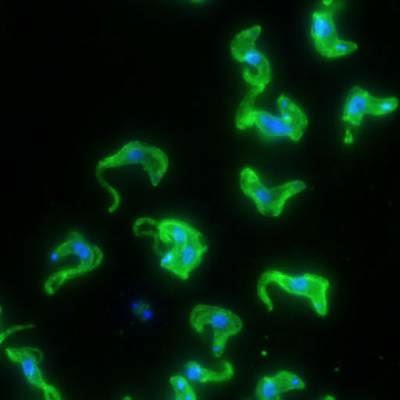
T. brucei‘s protein disguise comes from a family of variant surface glycoproteins (VSGs). At any time, a single VSG covers the parasite and, during an infection, the parasites in the bloodstream periodically switch to a new VSG. Together with Nina Papavasiliou and André and Bella Meyer Professor Emeritus George Cross, Monica Mugnier, a graduate student in Papavasiliou’s lab, set out to take a close look at these switches by tracking the VSG genes expressed over time during an infection.
“Little is known about the dynamics of VSG switching by the sleeping sickness parasite. What is known is that the coat is highly antigenic and stimulates a very strong immune response that wipes out most of the parasite population. The prevailing paradigm regarding the ability of the parasite to evade the immune response comes from studies completed decades ago, which suggested that a parasite changes its coat and stimulates an immune response on a weekly basis. Monica’s work totally upends this picture, and therefore the assumptions about how this parasite interacts with the immune response” says study author Associate Professor Nina Papavasiliou, head of the Laboratory of Lymphocyte Biology.
 A novel technique Mugnier developed to track the rise and fall of individual VSGs over time made this new insight possible. , “Previously, it was assumed that, as the immune system begins to recognize a parasites’ VSG and clear it, only one or a few VSGs would emerge to make up the next wave of parasites. However, our results contradict this paradigm,” Mugnier says. “We find as many as 80 distinct trypanosome coats at any given time”.
A novel technique Mugnier developed to track the rise and fall of individual VSGs over time made this new insight possible. , “Previously, it was assumed that, as the immune system begins to recognize a parasites’ VSG and clear it, only one or a few VSGs would emerge to make up the next wave of parasites. However, our results contradict this paradigm,” Mugnier says. “We find as many as 80 distinct trypanosome coats at any given time”.
“This has substantial implications for the interaction of this parasite with its host” says Cross. For example, it is now clear that the diversity of the VSGs required to maintain an extended infection exceeds the number of functional genes responsible for encoding them. This raised a question, since T. brucei infections can last for years. How does the parasite generate enough unique VSGs to continue fending off the immune system for that long, considering its repertoire of pre-existing genes?
Previous research suggested the answer: by mixing and matching. T. brucei has nearly 2,000 functioning and nonfunctioning VSG genes. By recombining these, the parasite can create many new disguises. This mechanism is likely a crucial source of VSG diversity for the parasite, according to the researchers, who identified a number of so-called mosaic VSGs, and, for the first time, documented the timing of their appearance during the course of the infection.
“Antigenic variation is both key to T. brucei‘s success and its Achilles’ heel. This strategy has made it impossible to develop a vaccine against sleep sickness, but without antigenic variation this parasite would not stand a chance against a healthy immune system,” Mugnier says. “The methods we developed lay the foundation for a better understanding of the diseases caused by African trypanosomes, and by other pathogens whose long-term infections are facilitated by antigenic variation, which could improve our strategies for fighting these infections.”
Image: Trypanosoma brucei, the African sleeping sickness parasite; Taken by Danae Schulz/Laboratory of Lymphocyte Biology/Rockefeller University